Labeling cortical sulci
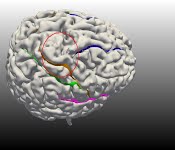
Sulci are fissures or grooves on the cortical surface. They serve as orientating landmarks
and also allow the neurosurgeon access to different parts of the brain. The major sulci
partition important functional areas.
Cortical sulci are highly variable. They vary in their physical description (shape, scale,
placement), in branching morphology and in their number. Sulci vary not just across
individuals but even between the hemispheres of a single brain. As much as we need a a
reliable automated labeling scheme to identify and label the sulci this variability poses a
challenge.
Labeling with multidimensional scaling
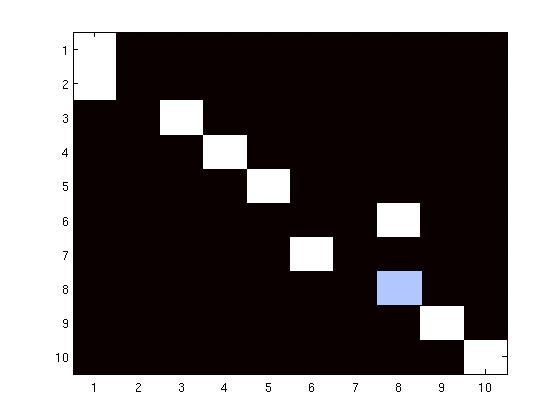
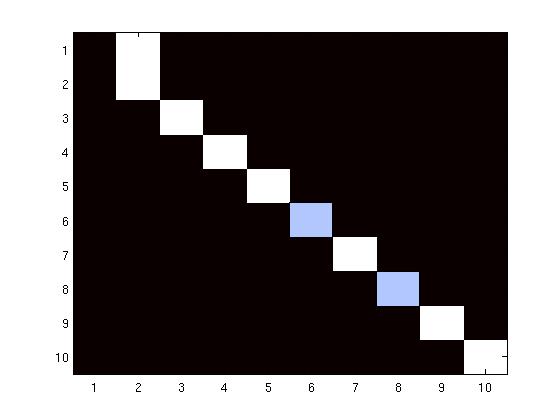
Multidimensional scaling (MDS) gives a physical map with which we can reproduce the
structural relationships between sulci in an embedded space. Unlabeled sulci, projected into
the same space using an out-of-sample procedure, are given class designations based on
nearest neighbor (NN) search. In experiments using a leave-one-out strategy, 90% of the 180
sulci drawn from 10 sulcal classes are successfully classified as are data from 5 out of 6
patients with cortical tumors.
With this supervised learning scheme, we offer a simple and intuitive solution to a
challenging problem. The use of relational matching gives us the flexibility to accommodate
normal variation in the sulci.
Publications
Mani, M., Srivastava, A., Barillot, C. The Labeling of Cortical Sulci using Multidimensional
Scaling , MICCAI Workshop on Manifold Learning in Medical Imaging (MMI 2008), New York City,
USA, September 2008.
[ Full paper ]
[ BibTex ]
[ Slides (pdf, 2.6M) ]
[ Poster ]
Mani, M., Supervised Labeling of Brain Sulci Using Multidimensional
Scaling.
[ Full paper ]
[ BibTex ]

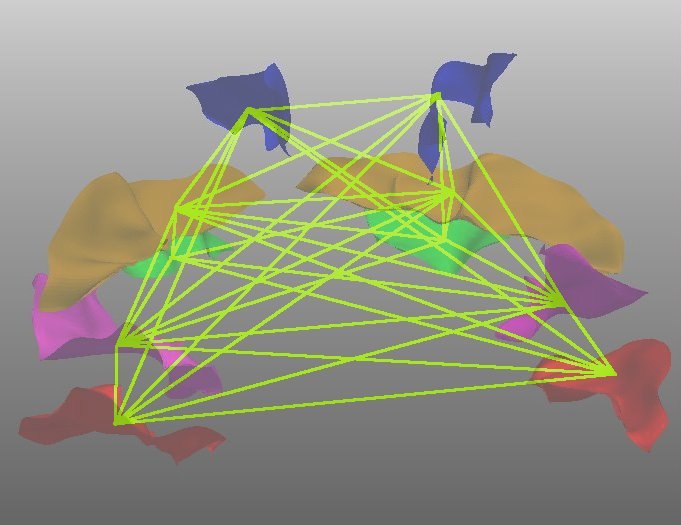
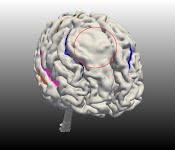
Labeling sulci of tumor subjects
The algorithm was tested on data from 6 subjects with medium to large tumors. The lesions
had displaced sulci or had otherwise altered the arrangement of the surrounding cortical
tissue. The sulci in the unaffected regions were successfully labeled, i.e. they had the
same error rate as for the healthy subjects. The value in our method lies in the fact that
we were able to classify sulci in the region surrounding the tumor in 5 out of 6 cases as
well.
The preliminary success with labeling tumor datasets suggests that we can design a
general-purpose tool to identify both normal and pathological sulcal data.